Chapter 11 Adding Papers and Slides to a Snakemake Workflow
11.1 Learning Objectives
TBD
!! TODO: warning on Rmarkdown / bookdown etc installs
11.2 State of Our Snakefile
MODELS = glob_wildcards("src/model-specs/{fname}.json").fname
# note this filter is only needed coz we are running an older version of the src files, will be updated soon
DATA_SUBSET = glob_wildcards("src/data-specs/{fname}.json").fname
DATA_SUBSET = list(filter(lambda x: x.startswith("subset"), DATA_SUBSET))
PLOTS = glob_wildcards("src/figures/{fname}.R").fname
TABLES = glob_wildcards("src/table-specs/{fname}.json").fname
##############################################
# TARGETS
##############################################
rule all:
input:
expand("out/figures/{iFigure}.pdf",
iFigure = PLOTS),
expand("out/tables/{iTable}.tex",
iTable = TABLES)
rule make_tables:
input:
expand("out/tables/{iTable}.tex",
iTable = TABLES)
rule run_models:
input:
expand("out/analysis/{iModel}.{iSubset}.rds",
iSubset = DATA_SUBSET,
iModel = MODELS)
rule make_figures:
input:
expand("out/figures/{iFigure}.pdf",
iFigure = PLOTS)
##############################################
# INTERMEDIATE RULES
##############################################
# table: build one table
rule table:
input:
script = "src/tables/regression_table.R",
spec = "src/table-specs/{iTable}.json",
models = expand("out/analysis/{iModel}.{iSubset}.rds",
iModel = MODELS,
iSubset = DATA_SUBSET),
output:
table = "out/tables/{iTable}.tex"
shell:
"Rscript {input.script} \
--spec {input.spec} \
--out {output.table}"
rule model:
input:
script = "src/analysis/estimate_ols_model.R",
data = "out/data/mrw_complete.csv",
model = "src/model-specs/{iModel}.json",
subset = "src/data-specs/{iSubset}.json"
output:
estimate = "out/analysis/{iModel}.{iSubset}.rds"
shell:
"Rscript {input.script} \
--data {input.data} \
--model {input.model} \
--subset {input.subset} \
--out {output.estimate}"
rule figure:
input:
script = "src/figures/{iFigure}.R",
data = "out/data/mrw_complete.csv",
subset = "src/data-specs/subset_intermediate.json"
output:
fig = "out/figures/{iFigure}.pdf"
shell:
"Rscript {input.script} \
--data {input.data} \
--subset {input.subset} \
--out {output.fig}"
rule gen_regression_vars:
input:
script = "src/data-management/gen_reg_vars.R",
data = "out/data/mrw_renamed.csv",
param = "src/data-specs/param_solow.json"
output:
data = "out/data/mrw_complete.csv"
shell:
"Rscript {input.script} \
--data {input.data} \
--param {input.param} \
--out {output.data}"
rule rename_vars:
input:
script = "src/data-management/rename_variables.R",
data = "src/data/mrw.dta"
output:
data = "out/data/mrw_renamed.csv"
shell:
"Rscript {input.script} \
--data {input.data} \
--out {output.data}"
##############################################
# CLEANING RULES
##############################################
rule clean:
shell:
"rm -rf out/*"
rule clean_data:
shell:
"rm -rf out/data/*"
rule clean_analysis:
shell:
"rm -rf out/analysis/*"11.3 Where We Are
In the last chapter we constructed a new target rule,
which we called all,
to combine the table and figure outputs.
This enabled us to be able to run the whole project from start to finish with the command snakemake --cores 1.
At this end point our workflow is illustrated by the following rulegraph:
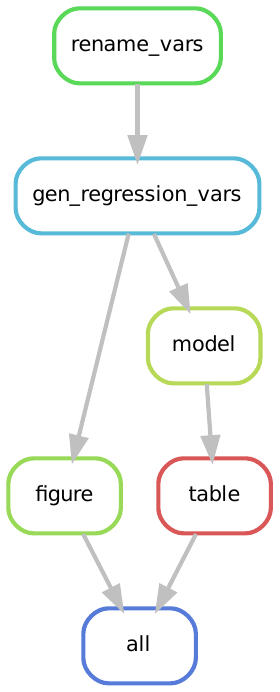
The possibility to end our workflow with the outputs of our data analysis is one possible end-point. An alternative endpoint is to extend the workflow to also manage the build of the literate outputs of the project, which in most cases is a paper and a set of slides. In what follows we explore this extension.
11.4 Building a Paper
Our project’s source files have everything we need to construct a paper:
- Data analytic outputs in the form of graphs and tables, in
out/figuresandout/tables - Text of a short paper, located in
src/paper/ - A script to construct the paper, located in
src/lib/build_article.R15
This suggests that a rule to build a paper might look like the following:
rule build_paper:
input:
script = "src/lib/build_article.R",
<SOME WAY TO ADD TABLES, FIGURES AND PAPER CONTENTS>
output:
pdf = "out/paper/paper.pdf"
shell:
"Rscript {input.script} <SOME_CLI_OPTIONS>"This is the structure we want to adopt.
To see how we can flesh out the remaining structure,
where <SOME...> suggests we need further specification,
let’s look at what the script src/lib/build_article.R expects as command line arguments:
which yields:
Usage: src/lib/build_article.R [options]
Options:
-i CHARACTER, --index=CHARACTER
Name of rmarkdown file where paper metadata is located
-h, --help
Show this help message and exit
This tells us we need to find an Rmarkdown file with the paper metadata.
Intuitively, the project’s structure tells us we should look for a file in src/paper.
Let’s do just that:
which returns the following:
01-intro.Rmd 03-data.Rmd _bookdown.yml _output.yml references.bib
02-literature.Rmd 04-results.Rmd index.Rmd preamble.texThere’s a lot there.
The file we are after is index.Rmd.
So let’s update our rule build_paper with what we have learned:
rule build_paper:
input:
script = "src/lib/build_article.R",
index = "src/paper/index.Rmd"
<SOME WAY TO ADD TABLES, FIGURES AND PAPER CONTENTS>
output:
pdf = "out/paper/paper.pdf"
shell:
"Rscript {input.script} --index {input.index}"Now, there is a lot of other files in src/paper/ apart from index.Rmd.
Here’s a brief summary of what they do:
01-intro.Rmd, …04-results.Rmdcontain the textual content of the paper, and insert the figures and tables where we need them. Each file is a section of the paper, and files are numbered in the order we want the sections to appear.16preamble.texcontains some additional latex packages and environments we need to build the articlereferences.bibcontains any references we will cite in the paper, in BIBTeX format.agsm.bstformats our bibliography._bookdown.yml- a file that tellsRwhere we want the output to be saved, and what to call it. We’ve set that up to be saved inout/paper/and the filename bepaper.pdf._output.yml- a file that tellsRhow to construct the paper, i.e. what template we need, and if we need other latex packages.
We need all these files to be inputs to the rule build_paper.
This is so a rebuild of the paper is triggered any time we change on of these files.
We could manually specify each one, or we could use the notion of glob_wildcards to collect the files by name,
and then use expand() to include all of these files in the rule.
First, let’s do the glob_wildcards step - we want all files in src/paper:
TABLES = ...
PAPER_FILES = glob_wildcards("src/paper/{fname}").fname
##############################################And then we update the rule build_paper once more to expand over all files we found in the glob_wildcards step:
rule build_paper:
input:
script = "src/lib/build_article.R",
paper = PAPER_FILES
<SOME WAY TO ADD TABLES AND FIGURES>
output:
pdf = "out/paper/paper.pdf"
shell:
"Rscript {input.script} --index src/paper/index.Rmd"Finally we need to add the figures and tables as inputs.
We’ll use the expand function to iterate over the FIGURES and and TABLES lists we previously created.
The build_paper rule is now:
rule build_paper:
input:
script = "src/lib/build_article.R",
paper = PAPER_FILES
figures = expand("out/figures/{iFigure}.pdf",
iFigure = PLOTS),
tables = expand("out/tables/{iTable}.tex",
iTable = TABLES)
output:
pdf = "out/paper/paper.pdf"
shell:
"Rscript {input.script} --index src/paper/index.Rmd"To build the rule with Snakemake:
At the conclusion of the run, we see that the file out/paper/paper.pdf is produced.
We also see that the tex file paper.tex is produced.
This can be useful when it’s time to submit the final version of paper to a journal after acceptance -
they most likely don’t want all our Rmds!.
11.5 Integrating build_paper output into all
Let’s compare the rules that need to be executed when we run
snakemake --cores 1 build_paper and snakemake --cores 1 all:
!! TODO add some space between, captions etc

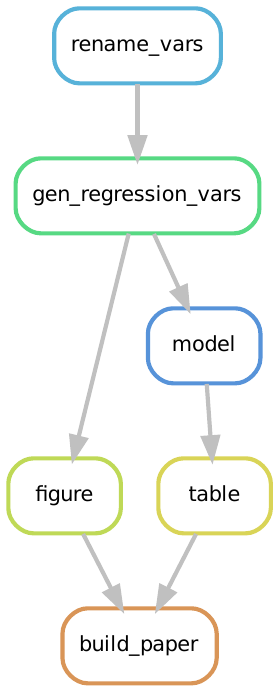
We see that they are the same.
The reason is that the build_paper rule has all of the figures and tables included.17
This suggests we can update the all rule.
We’ll update it by removing the figures and tables as inputs, and replacing them with the paper we produced:
Now when we execute Snakemake with snakemake --cores 1 all it will run the full workflow including the paper build.
That is, our rule graph becomes:
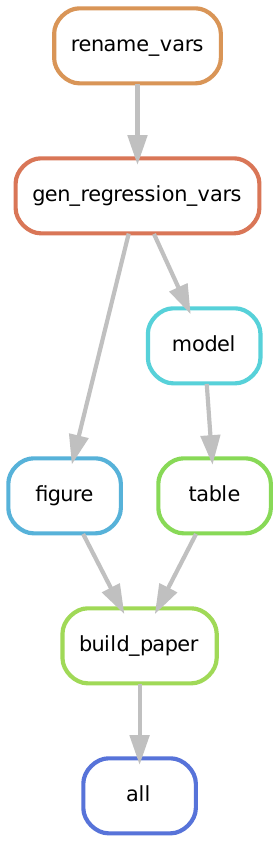
To see this in action, let’s clean our output directory:
!! TO DO include snippet from snakemake output.
The end OR beginning shows that the all rule now builds the paper after running all the data cleaning,
model estimations, and the figure & table creation.
!! TIP: What to do if paper doesn’t include all tables and figs?
Sometimes our final paper may not include all the tables and figures we have created. The question is then what do we want to do, and do we need to make all the additional tables and figures?
Here’s some thoughts on likely situations you could encounter and a possible solution:
If the additional tables and figures aren’t needed for the project, we don’t need to build them. Adjust the inputs to
build_paperto only include what is needed, and don’t build them all.If the additional tables and figures are needed, but they aren’t included in the final paper. This means they are likely going to be part of a long web appendix. Create a rule to build the web appendix, and then add the final web appendix doc as a input to the
allruleIf the additional tables and figures are needed, but they aren’t included in any of the final documents. One could leave the tables and figs as part of the
allrule in addition to the paper.
11.6 Making paper.pdf Easier to Find
So far we have used the all rule like any other target rule -
we specify a set of inputs we want to have built.
Our current all rule builds the final paper and any of its dependencies,
but there is nothing stopping us from extending this rule to work more for us, by taking the inputs
and doing something to them via shell command to create new outputs.
What we have in mind is taking the paper located in out/paper/paper.pdf and copying it to the project’s
root directory so it is easy to find.
Let’s do this by adding some additional structure to the all rule:
We want to create a copy of the paper, and put it in the root directory, so we can create the output as “paper.pdf.”18
And then we can copy from one directory to another using the cp command.
The first argument after cp is what we want to copy - in our case {input.paper},
and the last is where we want to copy the file to - which is {output.paper} for us.
Let’s make those changes:
rule all:
input:
paper = "out/paper/paper.pdf"
output:
paper = "paper.pdf"
shell:
"cp {input.paper} {output.paper}"Now run the all rule:
and then look at the contents of the project’s root directory:
find_r_packages.sh out/ README.md config.yaml
install_r_packages.R paper.pdf REQUIREMENTS.txt sandbox/ SnakefileAnd we can now see a copy of the paper located in the root directory, as desired.
Now everytime we run all, the latest copy of the paper will be easily available in the root direcroty.
11.6.1 Exercise: Adding a Slide Deck.
Create a rule
build_slidesthat usessrc/lib/build_slides.Rmdto produce a slide deck that is saved toout/slides/slides.pdfThe slide content is the in foldersrc/slides.Our slides only use one table (
table06.tex) and one figure (uncondtional convergence). Did you chose to include all figures and tables as inputs? Why or why not?Add the slide output to the
allrule, and make sure the all rule copies them across to the project’s root directory. (HINT: you can chain together copy commands in the shell with&&)
11.6.2 Exercise Solution
MODELS = glob_wildcards("src/model-specs/{fname}.json").fname
# note this filter is only needed coz we are running an older version of the src files, will be updated soon
DATA_SUBSET = glob_wildcards("src/data-specs/{fname}.json").fname
DATA_SUBSET = list(filter(lambda x: x.startswith("subset"), DATA_SUBSET))
PLOTS = glob_wildcards("src/figures/{fname}.R").fname
TABLES = glob_wildcards("src/table-specs/{fname}.json").fname
PAPER_FILES = glob_wildcards("src/paper/{fname}").fname
##############################################
# TARGETS
##############################################
rule all:
input:
paper = "out/paper/paper.pdf",
slides = "out/slides/slides.pdf"
output:
paper = "paper.pdf",
slides = "slides.pdf"
shell:
"cp {input.paper} {output.paper} && cp {input.slides} {output.slides}"
rule make_tables:
input:
expand("out/tables/{iTable}.tex",
iTable = TABLES)
rule run_models:
input:
expand("out/analysis/{iModel}.{iSubset}.rds",
iSubset = DATA_SUBSET,
iModel = MODELS)
rule make_figures:
input:
expand("out/figures/{iFigure}.pdf",
iFigure = PLOTS)
##############################################
# INTERMEDIATE RULES
##############################################
rule build_slides:
input:
script = "src/lib/build_slides.R",
index = "src/slides/slides.Rmd",
table = "out/tables/table_06.tex",
figure = "out/figures/unconditional_convergence.pdf",
preamble = "src/slides/preamble.tex"
output:
pdf = "out/slides/slides.pdf"
shell:
"Rscript {input.script} --index {input.index} --output {output.pdf}"
rule build_paper:
input:
script = "src/lib/build_article.R",
paper = expand("src/paper/{iPaper}",
iPaper = PAPER_FILES),
figures = expand("out/figures/{iFigure}.pdf",
iFigure = PLOTS),
tables = expand("out/tables/{iTable}.tex",
iTable = TABLES)
output:
pdf = "out/paper/paper.pdf"
shell:
"Rscript {input.script} --index src/paper/index.Rmd"
# table: build one table
rule table:
input:
script = "src/tables/regression_table.R",
spec = "src/table-specs/{iTable}.json",
models = expand("out/analysis/{iModel}.{iSubset}.rds",
iModel = MODELS,
iSubset = DATA_SUBSET),
output:
table = "out/tables/{iTable}.tex"
shell:
"Rscript {input.script} \
--spec {input.spec} \
--out {output.table}"
rule model:
input:
script = "src/analysis/estimate_ols_model.R",
data = "out/data/mrw_complete.csv",
model = "src/model-specs/{iModel}.json",
subset = "src/data-specs/{iSubset}.json"
output:
estimate = "out/analysis/{iModel}.{iSubset}.rds"
shell:
"Rscript {input.script} \
--data {input.data} \
--model {input.model} \
--subset {input.subset} \
--out {output.estimate}"
rule figure:
input:
script = "src/figures/{iFigure}.R",
data = "out/data/mrw_complete.csv",
subset = "src/data-specs/subset_intermediate.json"
output:
fig = "out/figures/{iFigure}.pdf"
shell:
"Rscript {input.script} \
--data {input.data} \
--subset {input.subset} \
--out {output.fig}"
rule gen_regression_vars:
input:
script = "src/data-management/gen_reg_vars.R",
data = "out/data/mrw_renamed.csv",
param = "src/data-specs/param_solow.json"
output:
data = "out/data/mrw_complete.csv"
shell:
"Rscript {input.script} \
--data {input.data} \
--param {input.param} \
--out {output.data}"
rule rename_vars:
input:
script = "src/data-management/rename_variables.R",
data = "src/data/mrw.dta"
output:
data = "out/data/mrw_renamed.csv"
shell:
"Rscript {input.script} \
--data {input.data} \
--out {output.data}"
##############################################
# CLEANING RULES
##############################################
rule clean:
shell:
"rm -rf out/*"11.7 Final Thoughts
!! TODO: Box titled: When to Extend the Workflow to Paper and Slides?
Note that the script to construct the paper is in
src/librather thansrc/paper. This is because the scriptbuild_article.Ris quite general and can easily be used across multiple projects. We like to put scripts that are not specific to the current project insrc/lib.↩︎By default,
bookdown- ourRpackage that builds the paper wants to insert files in an alpha-numeric order afterindex.Rmd. We go along with this by numbering files in the order we want them. There’s also a way to manually specify the order as a list of file names, but we think this numeric way is a bit easier. (It also keeps the files in order in our directory so we can glimpse the structure and easily find the file we want to edit.) ↩︎If we didn’t include all the tables and figures in the
build_paper, Snakemake would only build the ones required by that rule. What we say below depends on all tables and figures being included. We will have a remark on what to do if this is not the case in due course.↩︎Note that by not putting any directory before
paper.pdfit refers to the current directory (the project root) by default.↩︎