Chapter 3 Initial Steps with Snakemake
Overview
Questions
- Where and how do we write Snakemake rules?
- How do we execute a rule?
- How can rules interact with input and output files?
- How does Snakemake deal with redundant executions?
Objectives
- Recognize the key parts of a Snakemake rule.
- Run Snakemake from the shell.
- Explain when and why snakemake executes a rule.
- Understand how to read the output of
snakemake --summary.
Now that we’ve explored the structure of our project, we are ready to start building a fully reproducible pipeline.
Snakemake expects instructions in a file called Snakefile.
Typically we put the Snakefile in the project’s main folder (i.e. in ./).
The Snakefile contains a collection of rules that together define the order in which a project will be executed.
We have added an empty Snakefile in the main project folder.
You can edit this file in a text editor of your choice.
In the remainder of this tutorial we will edit the file together, gradually constructing the pipeline which reproduces the results from MRW.
3.1 Writing Our First Rule
All good software tutorial seem to start with a hello world example, so we’ll follow suit and do the same.
To do so, we open Snakefile in a text editor of our choice and write the following text into the file:
The first line in our example defines the name of a rule, in this case hello_world.
The part following shell: tells Snakemake that this rule executes a shell command, which in our case prints Hello World! to the console.
The indentation here is important.
We need the shell command to be indented in one TAB space from rule.
Additionally we need the shell command 'echo "Hello World!"' to be indented on TAB space further than shell.
Snakemake uses the indentation to figure out different components of the each rule.
Snakefiles are Written as Python 3 Code
Snakemake instructions are written in Python 3 with the addition of a a few extra objects and methods the Snakemake package provides. The structure of our rule will look very familiar to anybody who has written Python functions before. The fact that Snakemake is written in Python is why indentation matters. This implies that indentation is interpreted by the language and that deviations from correct indentation will result in errors.
Throughout the rest of the tutorial, those familiar with Python will continue to see similarities to Python code. If you do not know Python, there’s no need to worry - our explanations and what you need to follow along will not depend on it.
3.2 Executing a Rule
After saving our changes to Snakefile, we can execute the newly created rule by navigating to the project’s main folder and by typing Snakemake --cores 1 into the terminal and then hitting RETURN.
By default Snakemake will execute the first rule it encounters in a Snakefile.
As our file only contains a single rule, it will execute the hello_world rule and print Hello World! to the console together with some additional information about the execution of the work flow:
Building DAG of jobs...
Using shell: /usr/bin/bash
Provided cores: 1 (use --cores to define parallelism)
Rules claiming more threads will be scaled down.
Job counts:
count jobs
1 hello_world
1
[Fri Feb 26 18:11:00 2021]
rule hello_world:
jobid: 0
Hello World!
[Fri Feb 26 18:11:00 2021]
Finished job 0.
1 of 1 steps (100%) doneWhen a file contains more than a single rule, we often want to be more explicit and tell Snakemake to execute a particular rule like so:
In our case snakemake --cores 1 hello_world will give the same output as the last execution.
3.2 --cores
At this point you are probably guessing that the long option --cores SOME_NUMBER tells Snakemake how many CPU cores to use when executing a workflow.
This is correct, and snakemake --cores 1 tells Snakemake to use a single core of your computer when it runs.
Running workflows in parallel, by specifying for example snakemake --cores 4 can speed up complex workflows substantially when a workflow’s rules can be easily parallelized.
In this guide we will execute all workflows with a single core to make the code compatible with all computers.
You are encouraged play around with the core count yourself to and see what effect this may have.
3.2 Snakefiles do not have be be called Snakefile
We do not have to call our Snakefile Snakefile.
However, if we call it something else, we will need to tell Snakemake where to find the set of rules.
We can do this with the -s flag.
For example, if our Snakefile is names SomeSnakefile:
Sometimes the suffix .smk is used by researchers to identify Snakefiles not called Snakefile,
for example install.smk
3.3 First Steps in the MRW Pipeline
Like most empirical research, our project starts with data management and data cleaning steps.
The data management subdirectory, src/data-management contains two scripts:
rename_variables.R
gen_reg_vars.Rwhich need to be executed sequentially.
In this chapter, we want to run the script rename_variables.R to tidy up the variable names in our data set.
In the next chapter, we will then add gen_reg_vars.R, which creates additional variables to be used in subsequent steps.
3.3.1 Using the Rscript command to execute R scripts
We can run any R script with the Rscript shell command.
The command expects the path to an .R script as the first argument.
Simple R scripts that do not require additional options can be run as:
Most scripts in this project have been written to require additional parameters.
To see what additional parameters are needed, we can run the R script with the --help option.
Let’s apply this and execute the rename_variables.R script with the --help flag:
This prints the following output to the console:
Usage: src/data-management/rename_variables.R [options]
Options:
-d CHARACTER, --data=CHARACTER
stata dataset file name
-o CHARACTER, --out=CHARACTER
output file name [default = out.csv]
-h, --help
Show this help message and exitThis suggests the script expects
- a
--dataoption which defines the filepath to a Stata data set. This is the input dataset which the script transforms. For us, this will be the filesrc/data/mrw.dta - a
--outoption which defines the filepath the transformed data set will be saved. We will save the output to the fileout/data/mrw_renamed.csv
Let’s set up our first rule for the MRW pipeline using the structure we learned from the hello_world rule.
We will name the rule rename_vars and place it at the top of our Snakefile:
rule rename_vars:
shell:
"Rscript src/data-management/rename_variables.R \
--data src/data/mrw.dta \
--out out/data/mrw_renamed.csv"Note that \ in the shell command allows us to write the command over several lines for better readability.
They can be omitted when we write the command in a single line.
Now if we run Snakemake:
We see that Snakemake executes the script for us, printing output to screen and saving the output file as expected. Used this way, we can see that a Snakemake rule is an instruction on how to execute part of a project’s code. This is true, but it can be even more powerful than that.
3.3.2 The input and output arguments
Our current rename_vars rule was simple - it specified one shell command.
We can add some further structure to this rule by specifying input and output files in a different manner.
We are going to specify rules in the following format:
rule rule_name:
input:
input_name1 = "PATH/TO/input_one",
input_name2 = "PATH/TO/input_two"
output:
output_name1 = "PATH/TO/SAVE/output_one",
output_name2 = "PATH/TO/SAVE/output_two"
shell:
"HOW TO MIX IT ALL TOGETHER"In this format, a Snakemake rule can be thought of as the recipe that combines the inputs to produce the outputs.
We can think of the input as ingredients, the output as the finished dish and the shell part as the recipe itself.
Snakemake expects these components to be provided in a particular way so that it knows what to do with the information we provide.
Snakemake rules can have as many inputs and outputs as the rule needs to function.
Each input and each output are given names, for example input_name1 which takes the value to the file path and name of the input file.
It is important to wrap each of these paths into quotations, and to separate each of the multiple inputs and outputs with a comma, ,.
3.3.3 A rule to rename variables.
Let’s now use the input and output syntax to update the rename_vars rule from above.
The rule will use the rename_variables.R script as a first input.
Let us therefore rename the first input into script and replace the right hand side with the path to the rename_variables.R script which we have used before when we printed the script’s help function.
Our rename_vars rule should now look something like this:
rule rename_vars:
input:
script = "src/data-management/rename_variables.R",
input_name2 = "PATH/TO/input_two"
output:
output_name1 = "PATH/TO/SAVE/output_one",
output_name2 = "PATH/TO/SAVE/output_two"
shell:
"Rscript src/data-management/rename_variables.R \
--data src/data/mrw.dta \
--out out/data/mrw_renamed.csv"Next, we want to add any additional inputs and also specify any outputs that the file produces.
From the help output we know the script expects a Stata dataset as an additional input, which we can find in the src/data/ folder.
We rename the second input into data and replace the right hand side with the path to Mankiw, Romer, and Weil (1992)’s Stata dataset.
Finally, we replace the first output name with data and define an appropriate output file path as the right hand side.
This is the location our transformed data will be saved to.
After these steps, our rule should look something like this:
rule rename_vars:
input:
script = "src/data-management/rename_variables.R",
data = "src/data/mrw.dta"
output:
data = "out/data/mrw_renamed.csv"
shell:
"Rscript src/data-management/rename_variables.R \
--data src/data/mrw.dta \
--out out/data/mrw_renamed.csv"After saving we can execute our new rule via:
Snakemake will give us some console output informing us about the execution of our rule. A successful execution will be displayed through green text ending in
If the execution was successful, we can look into our output directory to see if anything has happened:
which yields
mrw_renamed.csvOur file has been created as we expected.
Looking at our rule, there is a bit of repetition.
In the shell section, we repeat the full filepath of all the files use before in the input and output sections.
Snakemake does not require us to write these paths twice.
We can refer to the inputs and outputs in the shell part of our rule via the names we assigned to them.
For example, we can refer to the script “src/data-management/rename_variables.R” as {input.script},
and the data which is the output as {output.data}
Putting it all together will give us the following rule:
rule rename_vars:
input:
script = "src/data-management/rename_variables.R",
data = "src/data/mrw.dta"
output:
data = "out/data/mrw_renamed.csv"
shell:
"Rscript {input.script} \
--data {input.data} \
--out {output.data}"Now let’s run the rule once more:
We see the following output:
Building DAG of jobs...
Nothing to be done.This is not an error, it simply says that Snakemake doesn’t need to run the rule again. It’s time to think about why that could be the case.
3.4 No Redundant Executions
To understand the status of the project as seen by Snakemake, we can use the summary option of Snakemake:
In our case the following output is printed to the console:
Building DAG of jobs...
output_file date rule version log-file(s) status plan
out/data/mrw_renamed.csv Thu Jan 10 20:31:08 2019 rename_vars - ok no update
Snakemake tells us the status of the output file of the rename_vars rule is ok and that it plans not to update the file.
Now, let us see what happens if we update one of the input files.
The following command updates the time that rename_variables.R was last edited without changing the actual content of the file:
Running the summary command again
yields the following output:
Building DAG of jobs...
output_file date rule version log-file(s) status plan
out/data/mrw_renamed.csv Thu Jan 10 20:31:08 2019 rename_vars - updated input files update pendingSnakemake has detected that one input file is more recent than the output file. It therefore will execute the rule when asked to do so.
Running the rule again via:
will execute the rule.
We see one major advantage of Snakemake over alternative ways to execute projects. Snakemake keeps track of updates to input files and dependencies between files.This allows Snakemake to only re-runs a rule if an input has changed. This means we no longer have to think about when we last ran a script and if we need to run it again. Snakemake’s management of dependencies will turn out to be quite advantageous as we develop a larger project. No longer do we need to think about order of execution and when to re-run files.8
3.5 Our Workflow in Graphs
Throughout Parts II to IV we are going to build up our workflow to reproduce MRW’s analysis pipeline. In our motivation chapter we explained how our final workflow should be a connection between a set of rules,which we have depicted graphically as:
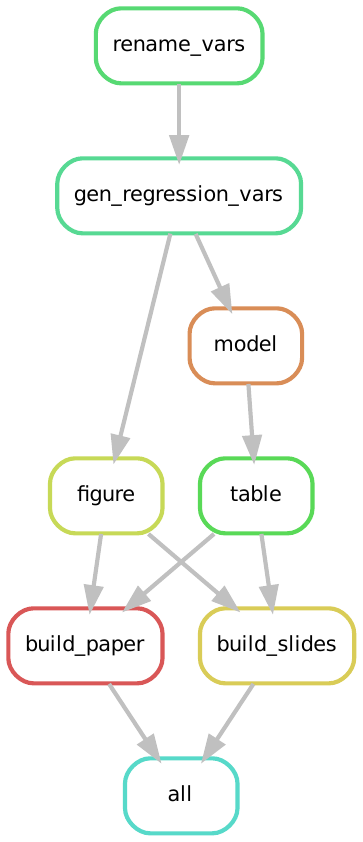
So far, we have constructed the first of those blocks an our set of rules in our current workflow looks like this:

We have made the first step in the data analysis pipeline. Let’s continue creating new steps in the next chapter.
B References
Mankiw, N. Gregory, David Romer, and David N. Weil. 1992. “A Contribution to the Empirics of Economic Growth*.” The Quarterly Journal of Economics 107 (2): 407–37. https://doi.org/10.2307/2118477.
Or, when to comment out chunks of a large script because “we don’t need to update that bit.”↩︎